Less is More: Use Case Driven Terminology Implementation
In healthcare, it is important to associate everything clinically relevant with a code to be able to accurately represent what you are observing or performing while providing patient care.
Surprisingly, including all the concepts in relevant clinical terminologies is counterproductive in certain use cases. For example, searching for the term “fever” in SNOMED CT returns 269 Disorders and 57 Findings. In a broader healthcare context, having the 327 options for representing the different kinds of fever may be relevant but if you are creating an application specifically to report COVID cases to public health you only need a few concepts.
Value sets and reference sets can be used to constrain the broader terminologies, similar to a view in a database. But using this strategy still requires the loading of large data sets into your applications which can affect the performance of applications that are built for a specific use case. For large enterprise applications, limiting the amount of terminology that is loaded and maintained may not be a significant consideration. However, smaller use case or lightweight applications will experience significant performance issues by loading complete versions of terminologies.
As an example, use cases dealing with just COVID related content require a much smaller working set of codes (while still retaining related content, full logical definitions, and other similar supporting content)
Full terminology concept counts vs COVID related concept counts
Benefits of Use Case Driven Terminology
Limits the universe for search/retrieval and data entry by humans
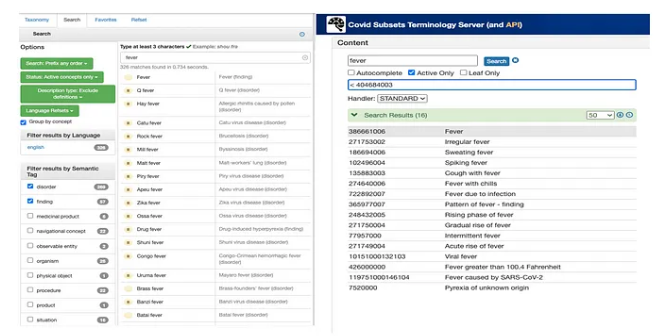
Fever Search Results: SNOMED Browser vs. COVID-19 Use Case Terminology Server
We have already discussed the benefits that can be realized by limiting the number of search results that are returned, as you can see in the screen shots of the fever example above.
Additionally, clinical terminologies like SNOMED CT can have multiple ways of representing the same data especially when paired with a statement model. For example, SNOMED has a single code for representing a COVID positive test, 897034005 |SARS-CoV-2 antibody test positive|. However, when using a statement model like HL7 you are most likely going to have a test that is performed for detecting SARS-CoV-2 and another field that would represent whether it was positive/detected, negative/not detected, or indeterminate.
By limiting the number of potentially irrelevant concepts, you can ensure that clinical data is more reliably and accurately captured, communicated, and retrieved.
2. Provide targeted sets of data in a single place for communication, research
Laboratories that were mandated to report COVID test results to public health authorities have a distinct set of values that are required for certain HL7 fields. During the pandemic, there were specialty laboratories that did not typically report results to public health authorities that were offering COVID testing. Having a terminology server available to provide just the terminology concepts that are needed to perform the reporting helps to speed up the creation of the messaging connections between the laboratories and the public health departments. For example, these message components only need a small set of data from terminologies for COVID reporting:
Test Ordered — 44 LOINC codes
Test Result — 11 SNOMED CT codes
Patient Demographics — small SNOMED CT Value sets
Specimen Source — 33 SNOMED CT codes
Ask at order entry (AOE) questions — Small subsets of LOINC, SNOMED CT, and HL7 codes
To support reporting COVID test results, only a few hundred codes are needed. This greatly simplifies the process of setting up a connection, reduces ambiguity as to which codes to use for discrete reporting, and ensures a more meaningful set of data upon which research can be done.
3. Public health monitoring and streamlined patient cohorting
Having a smaller, integrated set of codes that can be rapidly deployed to monitor public health messages would be useful for not only those entering data but also the public health departments doing the monitoring. If the data entry is streamlined to ensure those entering data are doing so with an approved set of codes and only varying when needed, it will make monitoring messages more reliable.
Patient cohorting refers to the grouping of patients exposed to or infected with the same pathogen in the same inpatient room/geographic area. Restricting the amount of data needed for processing will streamline the search and criteria to more effectively pinpoint the strategies needed to effectively coordinate care and treat these patients. For example:
Patients who had long COVID that were given/not given certain treatments
Age / sex related differences in outcomes / adverse events
Patients who have been vaccinated / not vaccinated
4. Rapid turn-around as content evolves
In pandemics, knowledge about the disease can change rapidly, as we saw in the early months of the COVID-19 pandemic. The clinical terminologies needed to record data in these early stages must be able to change and evolve rapidly. However, clinical terminologies have historically been slow to change with some terminologies only releasing once or twice a year.
In emergency cases like a pandemic, most content providers will release addendum or patches that can be used until the next release. In practice, it can still take months for this content to be added. For example, the condition “Long COVID” was first reported in Spring of 2020 but wasn’t officially added to SNOMED CT until July 2021. The condition of “COVID toes” is still not available in SNOMED CT as of this publication. Finally, once it does get released, it is up to consuming organizations to update their content (which most organizations DO NOT do immediately upon release). Having a smaller set of content will allow you to quickly identify anything that is missing, request the addition to the appropriate standard or add it to a local extension you maintain.
Additionally, as new and underutilized concepts begin to get more use, content errors are found that need to be corrected quickly to ensure accurate data entry and data retrieval. For example, most of the antibody negative concepts are subtypes of 442225006 |Negative measurement finding (finding)|. However, as illustrated in the image below, there are 4 SARS-CoV-2 concepts that are direct subtypes of the upper-level clinical finding concept and should be corrected to be in the same location as the other measurement findings. In our COVID-19 Use Case Terminology Server, the clinical finding concept only has 37 subtypes instead of the 157 subtypes in the full SNOMED CT release, making this error much easier to identify and correct.
4 SARS-CoV-2 concepts that belong in different sub-hierarchy
5. Collaboration by multiple organizations to deploy specialized content where it is underrepresented
All the previously mentioned benefits help different organizations to collaborate on only the content that is relevant to them. These smaller, specialized sets of data are easier to browse and search and ”hide” unnecessary content to allow easier development and QA on relevant content.
Example Implementation: covid.terminology.tools
We produced an example implementation of a Use Case Driven Terminology Server using COVID-19 as our example use case. You can compare the simplicity in browsing and searching on this domain versus full terminology browsers like the SNOMED CT Browser. To produce this implementation, we took the following steps:
A. Identified sources of COVID value sets/reference sets
The first step in producing a use case driven terminology server is to identify the concepts from the clinical terminologies you need for a successful implementation. In the case of COVID we chose to be as comprehensive as possible. Some of the sources we used to produce the example implementation came from:
▪ SNOMED International COVID related guidance
▪ LOINC SARS-CoV-2 and COVID-19 related LOINC terms
B. Created a release of the terminology server using the identified concepts
Using the identified value sets/reference sets we then extracted the appropriate subsets from the clinical terminologies and deployed them in our West Coast Informatics (WCI) Terminology Server product. For those simple terminologies that do not use other concepts to provide further definition, we only had to extract the codes that were identified in the value set/reference set. For more complex terminologies, like SNOMED CT, we had to also include concepts that were used to provide a logical definition. In the case of SNOMED CT, we used SNOMED International’s subontology extraction code that was developed to produce the International Patient Summary.
C. Evaluated the terminology server for any missing content
After deploying the clinical terminologies to the WCI Terminology Server, we then used the incorporated browser to evaluate the hierarchies for any missing content. For the most part we focused on upper-level hierarchies to make sure they were complete and not missing any important concepts. The subontology extraction code that we used to produce the subset of SNOMED CT caused a few concepts to be moved to the upper-level primitive concepts and we needed to adjust our starting concepts to include a few more grouper concepts to make the hierarchies cleaner and easier to navigate.
D. Created local content and/or submitted local content to terminology authors
There was some content that ended up missing that we identified as clinically relevant for our use case, for example “COVID toes”. In this case, we added it to an extension as well as submitted the content to the US National Release Center to hopefully add it to a future release of SNOMED CT.
Summary
Comprehensive terminology is important in the provision of healthcare, but not all use cases require entire terminologies to be present. Limiting the amount of terminology content loaded into an application based on its use case can have many benefits both on the data entry side and the retrieval and secondary use of that data.
Less terminology makes it easier to get the search results you are looking for and restricts the data entry to the concepts that are needed to meet certain reporting or interoperability requirements based on the statement model you are using.
Once data is entered, you can search and retrieve it faster and with less cognitive effort because the search space is smaller.
To learn more, try out our example implementation at https://covid.terminology.tools/.